Following our initial conversation via email, James Redfern and I met so that I could find out more about microbiology and discuss my project is greater depth.
Redfern gave me more information about the work carried out at MMU by the microbiology research team. As well as applying for funding and tenders, the team have a good relationship with the industries that require independent microbial testing. It is a field that has so many real-world applications and yet there is still so much left to understand and discover, which I feel makes it incredibly exciting territory.

This photo and the one above show the labs at MMU, the first one is mostly used by PhD students and the second one is a teaching lab for undergrads, maintained by the technicians.
In a general discussion I noted the following:
- Bacteria don’t have a nucleolus in their cells like human and plant cells, instead they have a ‘jumble’ of DNA. Viruses are different to bacteria and are not considered to be living because they have incomplete DNA, meaning they require something else (a host cell) in order to reproduce (although this point is up for debate).
- Because they contain their entire DNA, bacteria reproduce by splitting. E. coli cells split every 20 minutes so a significant culture can develop within a few hours.
- DNA is expressed in a string of A, T, G and Cs. The ribosome of the cell ‘reads’ the DNA code 3 letters at a time, and then uses this information to produce a protein (amino acid).
- When there is a change in the DNA (which could be just 1 letter) it results in a mutation.
- Even within a single species of bacteria there is still a lot of variation in the DNA (mutations). This can dramatically alter how it behaves and how it affects humans. Some strains of one species of bacteria could be harmful to humans while others could be benign or beneficial.
- When a bacteria encounters a new environmental condition such as the introduction of an antibiotic, if it is not killed it may mutate to help it live better in the new environment. This is what has led to antibiotic resistance.
- There are already fully antibiotic resistant strains of bacteria. I have heard a lot about antibiotic resistance in the news and as a criticism of factory farming practices. The microbiology community is debating whether the greater threat comes from agriculture or hospitals, but either way it is not an over-exaggeration to say we could be on the verge of a catastrophe for the human race.
- Silver and copper are antimicrobial materials but recently resistance has been observed here as well.
- Redfern suggests that an entirely new approach is required to combat antibiotic resistance, requiring microbiologists to engage in an an interdisciplinary conversation with public health and economic professionals (since antibiotic use in food production leads to lowered costs). It seems more and more that collaboration between disciplines is necessary for dealing with global issues.
- However, despite these dooms-day predictions, it is important to remember that the vast majority of microbes are benign or beneficial for humanity.


These two images show the special bins used to dispose of hazardous materials. The transparency of the materials and design was interesting to me and could provide a point of reference for the aesthetic development of my work.
Redfern showed me the DNA sequencing of a single Pseudomonas aeruginosa cell, which is a common multi-drug resistant pathogen that regularly causes infection in hospitals. The DNA was well over 6,700,000 letters long! And this is only one strain of this species of bacteria. At this level, there is a practically infinite amount of data that could provide source material for artistic projects.
With specific relation to my project, we discussed how I might collect environmental samples. Swabbing surfaces with a cotton bud might not allow me to recover microbes in great enough numbers to culture, since even surfaces that appear very smooth can have huge rifts and crevices when viewed under a microscope. The microbes sit in the rift and the swab passes straight over. A machine called a White Light Profilometer allows you to look closely at the surface of a material to see exactly how smooth it is. There is one of these in the MMU labs so there is potential for me to bring objects in to explore under it.

Pond water sample.
A way to take a sample from a flat surface (such as a wall or door) could be to use contact plates. These are like Petri dishes but they are filled all the way to the top with agar (rather than just a proportion of the depth). They are usually used to do a blanket check on surfaces that should be sterile. The operator simply ‘stamps’ the surface with them to pick the microbes up straight onto the plate. The disadvantage of this way of sampling is that it that it takes a generic sample across the whole plate. I may not be able to identify different microbes or the number of microbes, since they are mingled together and spread across the whole plate. However, this could be a good starting point for me to understand which surfaces do lend themselves to culturing from, and to test how inks and dyes affect growth.
Another simple experiment I could undertake in the lab is to carry out a serial dilution. For this, the stages are:
- Place an object/something to sample from (such as a paintbrush) into a saline solution and agitate it
- Inoculate a Petri dish with this solution (I also noted that plates are usually inoculated across 3 lines, so that the sample reduces each time, resulting in 1 line of solid growth, 1 line of broken growth, and 1 line of limited growth with single spots that can then be extracted and examined in isolation)
- Dilute the solution to a 1:10 ratio and inoculate another plate
- Carry out the above step 8 more times, so you have 10 plates in all
- The number of microbes are reduced 10-fold each time, so the final plate should have only a few spots of growth. It should then be possible to count the spots and apply the maths backwards to work out how many microbes were in the first plate (which will probably appear completely covered in microbes)
Doing a serial dilution test would make my exploration more precise than a contact plate by allowing me to see how many microbes were in my original sample, and I may also be able to identify what the microbes are by viewing them under a microscope (or sequencing their DNA).
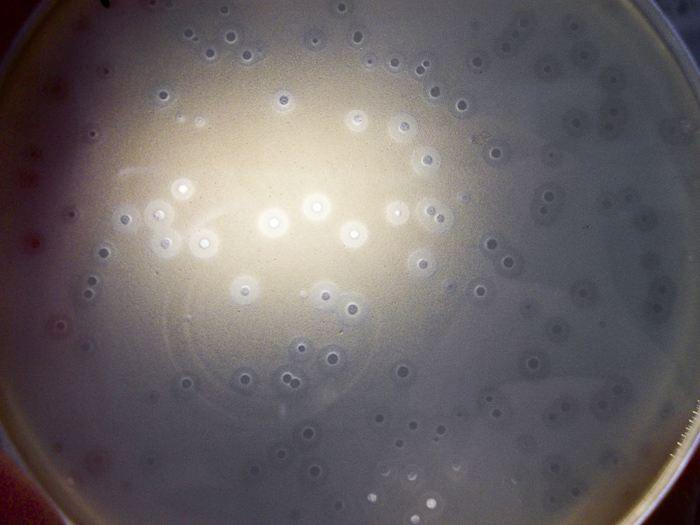
A Petri dish, held up to the light. The clear spots show a ‘kill zone’ where an antimicrobial substance has had effect. The halos around the clear spots are where the substance has had a partial effect.
Another interesting piece of information is the recent discovery that microbes behave differently in a community, which is known as a biofilm. When they get together, they can support each other, for example by passing water/nutrients from the top of the biofilm to the microbes at the bottom. They also communicate to each other. The microbes at the top may let the microbes at the bottom know to close their cell walls when they encounter an antibiotic. This research was pioneered by the molecular biologist Bonnie Bassler in 2002 (I intend to learn more about this from her TedTalk https://www.ted.com/speakers/bonnie_bassler)
The idea that the behaviour of bacteria is different when they are part of a unit (biofilm) has obvious correlation to the concerns and philosophies of Collaborative Practice and is highly interesting to me.
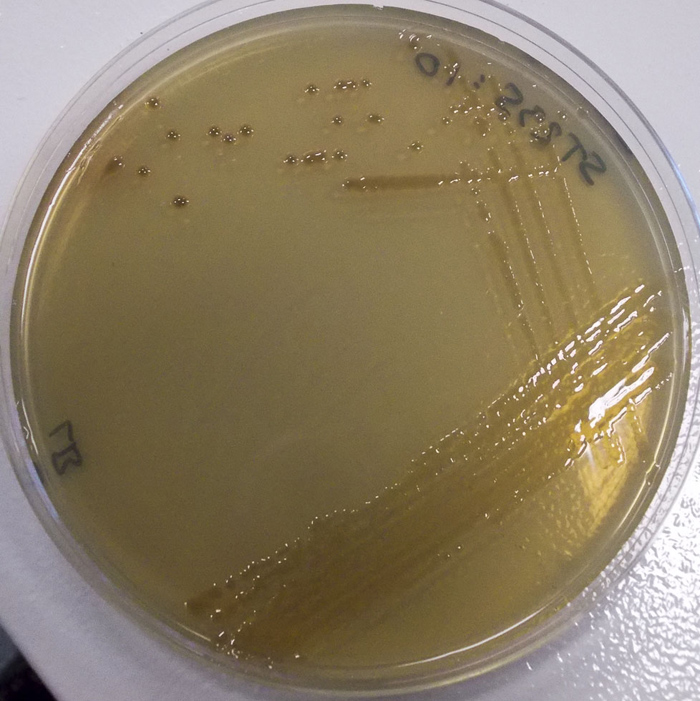
A plate that shows the ‘3-direction’ swabbing technique, used to reduce the amount of cultures in each direction. One of the single spots could be extracted for further examination.
Redfern also introduced me to a new key word: Microbiome. This is often used to refer to all of the microorganisms that live on and in a human subject, but it can also refer to the microbial profile of a place. In this project, I am studying the microbiome of my artist studio.
The next step is to consider how I may utilise the laboratory environment at MMU and complete a Risk Assessment for this activity.
Reference:
Redfern, J. (2017) Email correspondence and meeting. From 06 December 2016 – Ongoing. Subject: SciArt and Mould
